Summary
There are many different interactions existing between microorganisms and plants. Microorganisms are distinguished into pathogen and probiotics based on their behaviors with the host plant. The plant diseases are caused by pathogens, whereas probiotics trigger the plant defense and help the plant growth. One of the research interests in our lab is to study and reveal the mechanisms of interaction between pathogen/probiotics and plants. To address this research interest, the various approaches, such as big-data of bioinformatics, microbiology, biotechnology, and molecular biology, are being used. These findings provide novel information and knowledge for our lab as well as to the world, helping us to create innovative ideas like designing a new drug for plant diseases or novel biologics for promoting plant growth and defense responses.
微生物與植物之間有各種不同的相互作用。根據微生物對宿主植物的行為,其可被稱為病原或益生原。病原導致植物病害而益生原觸動植物防疫機制並促進植物生長。我們實驗室其中的研究興趣是探討病原/益生原與植物之間的相互作用。為了深入研究這個領域,我們利用了生物資訊的大數據、微生物學、生物科技及分子生物學等領域。這些研究發現對我們實驗室及世界都提供了新的資訊及知識,提供我們創新的想法,像是研發植物病害的新藥物或者製造新型生物製劑來促進植物生長及防禦反應。
Research
In our lab, we developed a ContigViews system that can help researchers to analyze next-generation sequencing (NGS) data and understand the mechanism of plant’s disease or defense through data mining. For example, bolting at high temperature in Lettuce (Lactuca sativa) is a big problem in the market. To understand the mechanism of heat-induced bolting in Lettuce, we performed transcriptomic analysis in ContigViews system (Fig.1). Through data mining, we found that Cyclic nucleotide-gated ion channels 4 (CNGC4) might play an important role in the heat-induced bolting.
We also investigate the regulatory role of small RNAs (sRNAs) in Arabidopsis thaliana, Solanum lycopersicum. Micro RNA (miRNA) and short interfering RNA (siRNA), which are 18 to 25 nucleotides (nt) non-coding RNAs, play important roles in controlling the gene expression of target mRNA in plants. In order to understand the overall regulatory networks of miRNA and siRNA, NGS is used. First, small RNA sequencing was performed to identify new microRNA (miRNA) and short interfering RNA (siRNA). Second, complementory base-pairing of small RNAs was performed by computer programming to predict potential target genes. Third, the predicted results were confirmed by degradome sequencing. These analyzed results will help scientists understand more about the regulatory networks of small RNAs in plants.
我們實驗室團隊研發了ContigViews 軟體。此軟體可協助研究人員分析次世代定序 (NGS) 及進行資料探勘,進而了解植物的疾病與防禦機制。例如,萵苣 (Lactuca sativa) 在高溫下抽苔是市場上的一個大問題。為了理解萵苣熱誘導抽苔的機制,我們透過 ContigViews 軟體進行轉錄組分析(圖1)。透過資料探勘,我們發現 Cyclic nucleotide-gated ion channels 4 (CNGC4) 可能在熱誘導抽苔中扮演了重要的角色。
我們也研究了小核糖核酸 (sRNAs) 在阿拉伯芥 (Arabidopsis thaliana, Solanum lycopersicum) 的調控角色。微核糖核酸 (miRNA) 和小干擾核糖核酸 (siRNA) 都是長18-25個核甘酸的非編碼核糖核酸,並且在控制植物目標 mRNA 的基因表現扮演重要的角色。我們透過 NGS,深入了解 miRNA 及 siRNA 的調控系統。首先,小片段的 RNA 序列用來辨認新的 miRNA 及 siRNA。第二,小 RNAs 的互補鹼基配對透過電腦程式來預測潛在的目標基因。第三,利用降解體定序來證實預測的結果。這些分析出來結果可以幫助科學家更加理解小 RNAs 在植物中的調控系統。
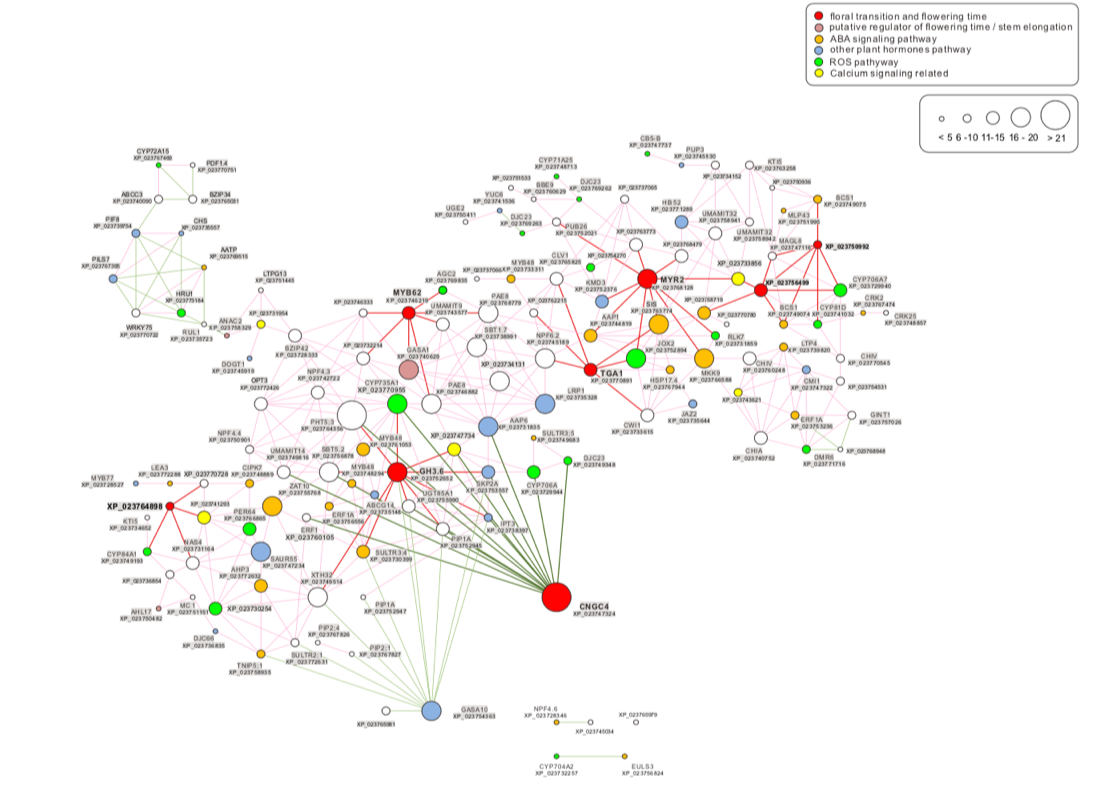
圖1基因-基因網絡。
To understand the function of P1 and HC-Pro of potyvirus in PTGS suppression. We generated many plasmids containing P1, HC-Pro and P1/HC genes of TuMV, ZYMV and TEV (Fig. A) and transgenic plants (Fig. B). The HC-Pro and P1/HC-Pro expression in plants induce serrated leaf phenotypes, but the P1/HCTu plant phenotype were stronger than HCTu plant, indicating HC-Pro alone have the ability to suppress plant PTGS and when P1 together with HC-Pro expresses in a plants can show enhanced HC-Pro-mediated PTGS suppression ability. In addition, we also found TuMV P1/HC-Pro specifically triggered AGO1 posttranslational degradation (Fig. C), but not in AGO2 (Fig. D). In the IP profiles, showed P1 proteins from TuMV, ZYMV and TEV viruses all can interact with VERNALIZATION INDEPENDENCE 3/ SUPERKILLER8 (VIP3/SKI8) protein. These findings can further understand how viral suppressor interfere plant gene silencing mechanism.
為了探討馬鈴薯Y病毒 (Potyvirus) 在 PTGS 抑制時,其 P1 及 HC-Pro 的功能。我們製造了許多質體,含有 P1,HC-Pro 及 P1/HC 基因的 TuMV, ZYMV, TEV (圖A) 及轉基因植物 (圖B)。植物中的 HC-Pro 和 P1/HC-Pro 表現會誘導葉子呈鋸齒狀,P1/HCTu 植物比 HCTu 植物的葉子呈更多鋸齒,這代表單單 HC-Pro 就有能力抑制植物PTGS,而 P1 及 HC-Pro 都在植物中表現顯示增強的 HC-Pro 介導 PTGS 的抑制能力。我們也發現 TuMV P1/HC-Pro 會專門誘發 AGO1 後轉譯降解 (圖C),AGO2 則不會 (圖D)。IP 位址顯示,來自TuMV, ZYMV 及TEV 病毒的 P1 蛋白質都與 VERNALIZATION INDEPENDENCE 3 / SUPERKILLLER 8 (VIP3/SKI8) 蛋白質相互作用。這些發現可用來更深入研究病毒抑制基因如何干擾植物基因靜默機制。
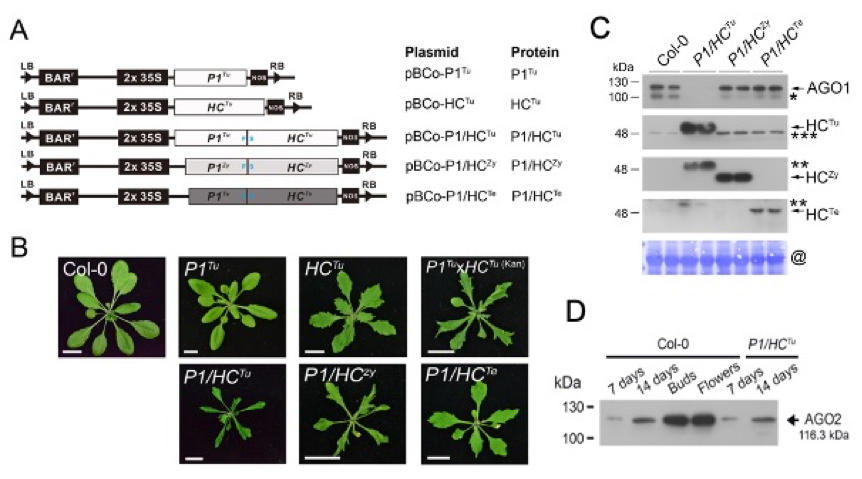
圖2 (A) 質體的示意圖。 (B) 轉基因植物中顯現不同程度的鋸齒狀葉子。在野生型與轉基因植物裡 (P1/HCTu, P1/HCZy 及 P1/HCTe) 內生的 AGO1
(C) 與 AGO2 (D) 蛋白偵測。
We investigated that how effectors of pathogens interfere gene silencing pathway. The PHYL1 effector of phytoplasma inhibits miR396 expression, resulting in mis-regulation of miR396 on the target mRNA SVP, which is an important negative flowering regulator. Mis-regulation on miR396-SVP mRNA causes leafy flower formation (Fig. 3). We thus created miR396 knockout plants through CRISPR/Cas9 method and miR396 overexpression transgenic Arabidopsis through T-DNA insertion to clarify whether miR396 could have impact on SVP expression level.
Besides, through bioinformatic prediction, we identified several potential proteins that interacted with PHYL1. In vivo immuno-precipitation experiments showed that IMP have physical interaction with PHYL1 among the candidates. However, no interaction was found during in vitro assay, implying other bridging proteins was involved in the interaction. Tracing back to the bioinformatic prediction, we considered some candidate proteins which might be the key connection of IMP and PHYL1 that it is still under investigation.
Meanwhile, we found that PHYL1 can interact with SEP3, the MADS transcription factor, to form a complex and caused MADS box degradation. We also found the amino acid sequence in a-helix of PHYL1 and SEP3_K were similar, implying their interaction (Fig. 4). Therefore, the interaction of SEP3_K and PHYL1 was examined by mutating the α-helix of PHYL1 and performing pull-down assays. The results showed that PHYL1 mutant did not interact with SEP3_K, proving that α-helix plays a critical role in PHYL1 and SEP3_K interaction.
我們探討病原的效應蛋白如何干擾基因靜默路徑。植物菌質體的 PHYL1 效應蛋白抑制 miR396 表現,導致 miR396 對於目標 mRNA SVP 調控失調,SVP 則是一個重要的負開花調控。miR396-SVP mRNA的失調導致葉化花形成 (圖3)。我們透過 CRISPR/Cas9 製造了剔除 miR396 基因的植物,也通過插入 T-DNA 使基轉阿拉伯芥 miR396 過度表現,確認 miR396 是否影響 SVP 表現程度。
此外,透過生物資訊的推測,我們已辨別幾個可能與 PHYL1 相互作用的蛋白質。活體免疫沉澱實驗顯示 IMP 蛋白與 PHYL1 有相互作用。但是,試管實驗並沒有顯示任何相互作用,這代表其他橋接蛋白也牽涉在這個作用中。再回推生物資訊的預測,我們發現並研究一些可能是 IMP 與 PHYL2 的橋接蛋白。另外,我們發現 PHYL1 會與 MADS 轉錄因子 SEP3 一起形成複合體並導致 MADS 盒子降解。我們也發現 PHYL1 與 SEP3_K 在α-螺旋有相似的氨基酸序列,圖4顯示他們的相互作用。因此,突變 PHYL1 的α-螺旋及進行拉下實驗可以檢驗 SEP3_K 與 PHYL1 之間的相互作用。實驗結果顯示突變的 PHYL1 沒有與 SEP3_K 互動,證明α-螺旋對 PHYL1 與 SEP3_K 之間的互動扮演重要的角色。

圖3 PHYL1 轉基因阿拉伯芥的葉化花。
圖3 PHYL1 轉基因阿拉伯芥的葉化花。
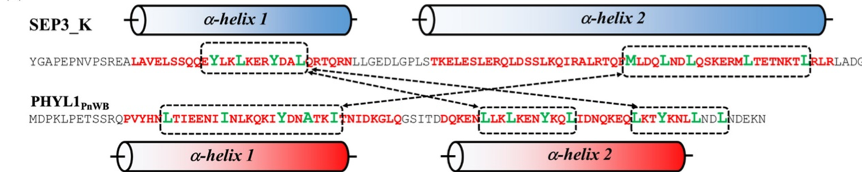
圖4 SEP3_K 與 PHYL1PnWB 相互作用的預測區域。
Marchantia polymorpha, also known as common liverwort, is the basal land plant lineage in a grade of bryophytes. This plant has been studied for more than 200 years to investigate plant evolution, life cycle, morphology and physiological responses. Now, it is considered as an excellent model plant because of the development of easy-to-use technical tools and with the establishment and release of genome sequences. To understand small RNA evolution in basal land plants, we identified MpMIR genes by comprehensive sequencing approaches and all of the information has been annotated in the genome database. In M. polymorpha, it is thought most of the miRNA species were novel and showed unique regulation, and only seven miRNA families have conserved targets in plants. We have cooperated with Laboratory of Plant Molecular Biology in Kyoto University for many years to perform CRISPR/Cas9 gene editing for genetic research (Fig. 5). We produced precise mutagenesis on MIRs and those core genes mediating RNA silencing pathway. Besides, we survey the MpAGO1-associated small RNA and try to figure out the characteristics of small RNA in M. polymorpha RISC complex. To better understand the roles of MpAGOs and two MpPIWI proteins on epigenetic regulation and phase transition, we also collected Mpago4, Mpago9, Mppiwia and Mppiwib mutants for further assay. We are happy to be in M. polymorpha research community and to share our findings to the worldwide.
Marchantia polymorpha 也就是地錢,屬於陸生植物中的地錢門。這個植物200多年前就被用來研究植物的演化、生命週期、型態及生理反應。如今,地錢是一個非常好的模式植物,其研究工具技術亦有一定發展及基因體序列也已完成。為了瞭解小核糖核酸在地錢門植物的演化,我們透過全面的定序識別了 MpMIR 基因,此基因的所有資料也已含在基因資料庫裡。M. polymorpha 大部分的 miRNA 都被認為是新種類且展示獨特的調控,只有7種 miRNA 家族在植物中有保守的目標。我們也與京都大學植物分子生物實驗室有多年的合作,利用 CRISPR/Cas9 進行基因編輯研究 (圖5)。我們製造出 MIRs 上準確的突變與調解 RNA 靜默途徑的主要基因。此外,我們調查有關 MpAGO1 的小核糖核酸並嘗試找出小核糖核酸在 M. polymorpha RISC 複合體的特質。為了更理解 MpAGOs 與兩個 MpPIWI 蛋白在表觀遺傳調控與轉變過程扮演的角色,我們收集了MpAGO4, MpAGO9, Mppiwia 及 Mppiwib 的突變來進行更多測試。我們非常高興可以在地錢的研究團隊中及向全球分享我們的發現。
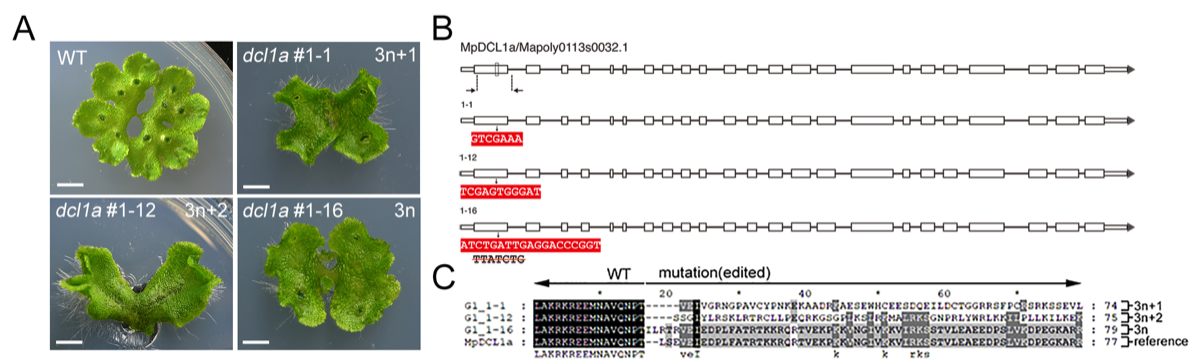
圖5 CRISPR/Cas9 介導的突變。(A) 不同表現型的 DCL1a 突變株與野生型生長在 1/2B5 培養基。(B,C) CRISPR/Cas9 產生的核苷酸與胺基酸序列插入與刪除的示意圖。
Publications
- Pan, Z.J., Wei, W.L., Tran, P.A., Fang, R.Y., Pham, T.H., Bowman, J.L., Chung, C.T., Shen, B.N., Yang, J.T., Chang, H.H., Jane, W.N., Cheng, C.H., Wang, C.C., Wu, H.Y., Hong, S.F., Shang, Q.W., Hu, S.F., Lin, P.C., Wu, F.H., Lin, C.S., Hung, Y.L., Shen, T.L., Lin, S.S.* 2025. HC-Pro inhibits HEN1 methyltransferase activity, leading to autophagic degradation of AGO1. Nature Communications 16:2503 (*Correspond author)
- Hong, S.F., Wei, W.L., Pan, Z.J., Cheng, S., Hung, Y.L., Tjita, V., Wang, H.C., Komatsu, A., Nishihama, R., Kohchi, T., Lo, J.C., Chiu, Y.H., Yang, H. C., Lu, M.Y., and Lin, S.S.. 2024. S insights into MpAGO1 and its regulatory miRNA, miR11707, in the high-temperature acclimation of Marchantia polymorpha. Plant Cell Physiol. (Revision) (*Correspond author)
- Liu, C.Y., Cheng, H.P., Lin, C.P., Liao, Y.T., Ko, T.P., Lin, S.J., Lin, S.S.*, and Wang, H.C*. 2024. Structural insights into the molecular mechanism of phytoplasma immunodominant membrane protein (IMP). IUCrJ. (Accepted) (*Co-correspond author)
- Hung, Y.L., Hong, S.F., Wei, W.L., Cheng, S., Yu, J.Z., Tjita, V., Yong, Q.Y., Nishihama, R., Kohchi, T., Bowman, J., Chien, Y.C., Chiu, Y.H., Yang, H.C., Lu, M.Y.J., Pan, Z.J.*, Wang, C.N.*, Lin, S.S.* 2024. Dual regulation of cytochrome P450 gene expression by two distinct small RNAs, a novel tasiRNA and miRNA, in Marchantia polymorpha. Plant Cell Physiol. (Accepted) (*Co-correspond author)
- Pan, Z.J., Wei, W.L., Tran, P.A., Fang, R.Y., Pham, T.H., Bowman, J.L., Chung, C.T., Shen, B.N., Yang, J.T., Hong, S.F., Shang, Q.W., Hu, S.F., Lin, P.C., Wu, F.H., Lin, C.S., Shen, T.L., Lin, S.S.* 2024. Turnip mosaic virus HC-Pro suppresses RNA silencing by sequencing HEN1 activity and enhancing autophagic AGO1 degradation. Nature Communications (Major revision) (*Correspond author) https://doi.org/10.21203/rs.3.rs-2131027/v1
- Kent, T.Y., de Jesús Castillo-Corea, B.R., Kumar, R., Lai, P.H., Lin, S.S., Wang, H.C. 2023. Shrimp SIRT4 promotes white spot syndrome virus replication. Fish & Shellfish Immunology. 109328
- Lee, Y.K., Lin, B.Y., Weng, T.H., Huang, C.K., Liu, C., Liu, C.C., Lin, S.S., Wang, H.C. 2023. Counting and measuring the size and stomach fullness levels for an intelligent shrimp farming system. Connection Science 35:2268878.
- Hong, S.F., Fang, R.Y., Wei, W.L., Jirawitchalert, S., Pan, Z.J., Hung, Y.L., Pham, T.H., Chiu, Y.H., Shen, T.L., Huang, C.K.*, and Lin, S.S.* 2023. Development of an assay system for the analysis of host RISC activity in the presence of a potyvirus RNA silencing suppressor, HC-Pro. Virology Journal 20:10 (*Co-correspond author)
- Chen, Y.R., Li, D.W., Wang, H.P., Lin, S.S., Yang, E.C. 2022. The impact of thigmotaxis deprivation on the development of the German cockroach (Blattella germanica). iScience 25:104802
- Bowman, J.L., Arteaga-Vazquez, M., Berger, F., Briginshaw, L.N., Carella, P., Aguilar-Cruz, A., Davies, K.M., Dierschke, T., Dolan, L., Dorantes-Acosta, A.E., Fisher, T.J., Flores-Sandoval, E., Futagami, K., Ishizaki, K., Jibran, R., Kanazawa, T., Kato, H., Kohchi, T., Levins, J., Lin, S.S., Nakagami, H., Nishihama, R., Romani, F., Schornack, S., Tanizawa, Y., Tsuzuki, M., Ueda, T., Watanabe, Y., Yamato, K.T., Zachgo, S. 2022. The Renaissance and Enlightenment of Marchantia as a model system. Plant Cell 34:3512-3542
- Lin, C.C., Lin, S.S., Chen, T.C. 2022. Complete genome sequence of Amazon lily mosaic virus isolated from amaryllis (Hippeastrum hybridum Hort.). Arch. Viol. 167:1495-1498
- Chiu, Y.‐H.; Hung, Y.‐L.; Wang, H.‐P.; Wei, W.‐L.; Shang, Q.‐W.; Pham, T.H.; Huang, C.‐K.; Pan, Z.‐J.*; Lin, S.‐S.* 2021. Investigation of P1/HC‐Pro‐mediated ABA/calcium signaling responses via gene silencing through high‐ and low‐throughput RNA‐seq approaches. viruses 13: 2349. (*co-correspond author)
- Sanobar, N., Lin, P.C., Pan, Z.J., Fang, R.Y., Tjita, V., Chen, F.F., Wang, H.C., Tsai, H.L., Wu, S.H., Shen, T.L., Chen, Y.H., Lin, S.S.* 2021. Investigating the viral suppressor HC-Pro inhibiting small RNA methylation through functional comparison of HEN1 in angiosperm and bryophyte. viruses 13:1837 (*correspond author)
- Huang, Y.H., Kumar, R., Liu, C.H., Lin, S.S., Wang, H.C. 2021. A novel C-type lectin LvCTL 4.2 has antibacterial activity but facilitates WSSV infection in shrimp (L. vannamei). Dev. Comp. Immunol. 126:104239.
- Chen, Y.R., Wei, W.L., Tzeng, D.T.W., Owens, A.C.S., Tang, H.C., Wu, C.S., Lin, S.S., Zhong, S., Yang, E.C. 2021. Effects of artificial light at night (ALAN) on gene expression of Aquatica ficta firefly larvae. Environ. Pollut. 281:116944
- Kumar, R., Tung, T.C., Ng, T.H., Chang, C.C., Chen, Y.M., Lin, S.S., Wang, H.C. 2021. Metabolic alterations in shrimp stomach during acute hepatopancreatic necrosis disease and effects of taurocholate on Vibrio parahaemolyticus. Front. Microbiol. 12:631468
- Chung, H.H., Ting, H.M., Wang, W.H., Chao, Y.T., Sung, Y.C., Hsieh, C.H., Lin, S.S., Hwu, F.Y., Shyur, L.F. 2020. Elucidation of enzymes involved in the biosynthetic pathway of bioactive polyacetylenes in Bidens pilosa using integrated omics approaches. J. Exp. Bot. 72:525-541
- Cheng, A.P., Chen, S.Y., Lai, M.H., Wu, D.H., Lin, S.S., Chen, C.Y., Chung, C.L. 2020. Transcriptome analysis of early defenses in rice against Fusarium fujikuroi. Rice. 13:65.
- Han, S., Ferelli, A.M., Lin, S.S., Micallef, S.A. 2020. Stress response, amino acid biosynthesis and pathogenesis genes expressed in Salmonella enterica colonizing tomato shoot and root surfaces. Heliyon 6:e04952.
- Hu, S.F., Wei, W.L., Hong, S.F., Fang, R.Y., Wu, H.Y., Lin, P.C., Sanobar, N., Wang, H.P., Sulistio, M., Wu, C.T., Lo, H.F., Lin, S.S*. 2020. Investigation of the effects of P1 on HC-Pro-mediated gene silencing suppression through genetics and omics approaches. Botanical Studies 61:22 (*correspond author)
- Huang, C.H., Foo, F.H., Raja, J.A., Tan, Y.R., Lin, T.T., Lin, S.S., Yeh, S.D. 2020. A conserved helix in C-terminal region of watermelon silver mottle virus NSs protein is imperative for protein stability affecting self-interaction, RNA silencing suppression and pathogenicity. Mol. Plant Microbe Interact. 33:637-652
- Apitanyasai, K., Chang, C.C., Ng, T.H., Ng, Y.S., Liou, J.H., Lo, C.F., Lin, S.S., Wang, H.C. 2020. Penaeus vannamei serine proteinase inhibitor 7 (LvSerpin7) acts as an immune brake by regulating the proPO system in AHPND-affected shrimp. Developmental and comparative immunology. 106:103600
- Montgomery, S.A., Tanizawa, Y., Galik1, B., Wang, N., Ito, T., Mochizuki, T., Akimcheva, S., Bowman, J., Cognat, V., Drouard, L., Ekker, H., Houng, S.F, Kohchi, T., Lin, S.S., Liu, L.Y.D., Nakamura, Y., Valeeva, L.R., Shakirov, E.V., Shippen, D.E., Wei, W.L., Yagura, M., Yamaoka, S. Yamato, K.T., Liu, C., Berger, F. 2020. Chromatin organization in early land plants reveals an ancestral association between H3K27me3, transposons, and constitutive heterochromatin. Current Biology 30:573-588
- Wen, C.H., Hong, S.F., Hu, S.F., Lin, S.S., Chu, F.H. 2020. Lfo-miR164b and LfNAC1 as Autumn Leaf Senescence Regulators in Formosan Sweet Gum (Liquidambar formosana Hance). Plant Science 291:110325
- Shelomi, M., Lin, S. S., Liu, L.Y. 2019. Transcriptome and microbiome of coconut rhinoceros beetle (Oryctes rhinoceros) larvae. BMC Genomics. 20:957
- Apitanyasai, K., Huang, S.W., Ng, T.H., He, S.T., Huang, Y.H., Chiu, S.P., Tseng, K.C., Lin, S.S., Chang, W.C., Bladwin-Brown, J.G. Long, A.D., Lo, C.F., Yu, H.T., Wang, H.C. 2019. The gene structure and hypervariability of the complete Penaeus monodon Dscam gene. Scientific Reports. 9:16595.
- Kumar, R. Ng, T.H., Chang, C.C., Tung, T.C., Lin S.S., Lo. C.F., Wang, H.C. 2019. Bile acid and bile acid transporters are involved in the pathogenesis of acute hepatopancreatic necrosis disease in white shrimp Litopenaeus vannamei. Research Cellular Microbiology. 22:e13127
- Liao, Y.T.*, Lin, S.S.*, Lin, S.J., Shen, B.N., Cheng, H.P., Lin, C.P., Ko, T.P., Chen, Y.F., Sun, W.T., Wang, H.C. 2019. Structural insights into the interaction between phytoplasmal effector causing phyllody 1 (PHYL1) and MADS transcription factor. Plant Journal 100:706-719 (*co-first author)
- Lo, K.J., Lin, S.S., Lu, C.W., Kuo, C.H., Liu, C.T. 2018. Whole-genome sequencing and comparative analysis of two plant-associated strains of Rhodopseudomonas palustris (PS3 and YSC3). Scientific Reports 8:12769
- Chen, Y.H., Shyu, Y.T.*, Lin, S.S.* 2018. Characterization of candidate genes involved in halotolerance using high-throughput omics in the halotolerant bacterium Virgibacillus chiguensis. PLOS ONE 13(8):e0201346 (*co-correspond author)
- Lin, S.S.*, Bowman, J.L. 2018. MicroRNAs in Marchantia polymorpha. New Phytologist. 220:409-416 (*correspond author)
- Ng, T.H., Lu, C.W., Lin, S.S., Chang, C.C., Tran, L.H., Chang, W.C., Lo, C.F., Wang, H.C. 2018. The Rho signaling pathway mediates the pathogenicity of AHPND-causing V. parahaemolyticus in shrimp. Cellular Microbiology 6:e12849
- Cheng, C.H., Shen, B.N., Shang, Q.W., Liu, L.Y.D., Peng, K.C., Chen, Y.H., Chen, F.F., Hu, S.F., Wang, Y.T., Wang, H.C., Wu, H.Y., Lo, C.T.*, Lin, S.S.* 2018. Gene-to-gene network analysis of the mediation of plant innate immunity by the eliciting plant response-like 1 (Epl1) elicitor of Trichoderma Formosa. Mol. Plant Microbe Interact. 31:683-691 (*co-correspond author)
- Flores-Sandoval, E., Eklund, D. M., Hong, S.F., Alvarez, J.P., Fisher, T.J., Lampugnani, E.R., Golz, J.F., Vázquez-Lobo, A., Dierschke, T., Lin, S.S., Bowman, J.L. 2018. Class C ARFs evolved before the origin of land plants and antagonize differentiation and developmental transitions in Marchantia polymorpha. New Phytologist 218:1612-1630
- Chen, Y.H., Lu, C.W., Shyu, Y.T.*, Lin, S.S.* 2017. Revealing the saline adaptation strategies of the halophilic bacterium Halomonas beimenensis through high-throughput omics and transposon mutagenesis approaches. Sci. Rep. 7:13037. (*co-correspond author)
- Huang, J.Y., Kang, S.T., Chen, I.T., Chang, L.K., Lin, S.S., Kou, G.H., Chu, C.Y., Lo, C.F. 2017. Shrimp miR-10a is co-opted by White spot syndrome virus to increase viral gene expression and viral replication. Front. Immunol. 8:1084
- Bowman, J.L., Kohchi, T., Yamato, K.T., Jenkins, J., Shu, S., Ishizaki, K., Yamaoka, S., Nishihama, R., Nakamura, Y., Berger, F., Adam, C., Aki, S.S., Althoff, F., Araki, T., Arteaga-Vazquez, M.A., Balasubrmanian, S., Barry, K., Bauer, D., Boehm, C.R., Briginshaw, L., Caballero-Perez, J., Catarino, B., Chen, F., Chiyoda, S., Chovatia, M., Davies, K.M., Delmans, M., Demura, T., Dierschke, T., Dolan, L., Dorantes-Acosta, A.E., Eklund, D.M., Florent, S.N., Flores-Sandoval, E., Fujiyama, A., Fukuzawa, H., Galik, B., Grimanelli, D., Grimwood, J., Grossniklaus, U., Hamada, T., Haseloff, J., Hetherington, A.J., Higo, A., Hirakawa, Y., Hundley, H.N., Ikeda, Y., Inoue, K., Inoue, S.I., Ishida, S., Jia, Q., Kakita, M., Kanazawa, T., Kawai, Y., Kawashima, T., Kennedy, M., Kinose, K., Kinoshita, T., Kohara, Y., Koide, E., Komatsu, K., Kopischke, S., Kubo, M., Kyozuka, J., Lagercrantz, U., Lin, S.S., Lindquist, E., Lipzen, A.M., Lu, C.W., De Luna, E., Martienssen, R.A., Minamino, N., Mizutani, M., Mizutani, M., Mochizuki, N., Monte, I., Mosher, R., Nagasaki, H., Nakagami, H., Naramoto, S., Nishitani, K., Ohtani, M., Okamoto, T., Okumura, M., Phillips, J., Pollak, B., Reinders, A., Rövekamp, M., Sano, R., Sawa, S., Schmid, M.W., Shirakawa, M., Solano, R., Spunde, A., Suetsugu, N., Sugano, S., Sugiyama, A., Sun, R., Suzuki, Y., Takenaka, M., Takezawa, D., Tomogane, H., Tsuzuki, M., Ueda, T., Umeda, M., Ward, J.M., Watanabe, Y., Yazaki, K., Yokoyama, R., Yoshitake, Y., Yotsui, I., Zachgo, S., Schmutz. J. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 171:287-304.
- Chou, W.C.*, Lin, S.S.*, Yeh, S.D.*, Li, S.L., Peng, Y.C., Fan, Y.H., Chen, T.C. 2017. Characterization of the genome of a phylogenetically distinct tospovirus and its interactions with the local lesion-induced host Chenopodium quinoa by whole-transcriptome analyses. PLoS One 12:e0182425. (*co-first author)
- Kanno, T., Lin, W.D., Fu, J.L., Wu, M.T., Yang, H.W., Lin, S.S., Matzke, A.J.M., Matzke, M. 2016. Identification of coilin mutants in a screen for enhanced expression of an alternatively spliced GFP reporter gene in Arabidopsis thaliana. Genetics 203:1709-1720
- Lin, P.C., Chang, Y.C., Lin, S.S.* 2016. An in vitro transcription/translation system for detection of protein interaction. Bio-protocol. 6:e1800. (*correspond author)
- Lin, P.C., Lu, C.W., Shen, B.N., Lee, G.Z., Bowman, J.L., Arteaga-Vazquez, M.A., Liu, L.Y.D., Hong, S.F., Lo, C.F., Su, G.M., Kohchi, T., Ishizaki, K., Zachgo, S., Althoff, F., Takenaka, M., Yamato, K.T., Lin, S.S.* 2016. Identification of miRNAs and their targets in the liverwort Marchantia polymorpha by integrating RNA-Seq and degradome analyses. Plant Cell Physiol. 57:339-358 (*correspond author)
- Bowman, J.L., Araki, T., Arteaga-Vazquez, M.A., Berger, F., Dolan, L., Haseloff, J., Ishizaki, K., Kyozuka, J., Lin, S.S., Nagasaki, H., Nakagami, H., Nakajima, K., Nakamura, Y., Ohashi-Ito, K., Sawa, S., Shimamura, M., Solano, R., Tsukaya, H., Ueda, T., Watanabe, Y., Yamato, K.T., Zachgo, S., Kohchi, T. 2015. The naming of names: guidelines for gene nomenclature in Marchantia. Plant Cell Physiol. 57:257-261.
- Chen, K.I., Pan, C.Y., Li, K.H., Huang, Y.C., Lu, C.W., Tang, C.Y., Su, Y.W.,Tseng, L.W., Tseng, K.C., Lin, C.Y., Chen, C.D., Lin, S.S.*, Chen, Y.T*. 2015. Isolation and identification of post-transcriptional gene silencing-related micro-RNAs by functionalized silicon nanowire field-effect transistor. Sci. Rep. 5, 17375 (*co-correspond author)
- Liu, C.T., Huang, H.M., Hong, S.F., Kuo-Huang, L.L., Yang, C.Y., Lin, Y.Y., Lin, C.P.*, Lin, S.S.* 2015. Peanut witches’ broom (PnWB) phytoplasma-mediated leafy flower symptoms and abnormal vascular bundles development. Plant Signal Behav. 10:e1107690 (*co-correspond author)
- Lee, C.T., Chen, I.T., Yang, Y.T., Ko, T.P., Huang, Y.T., Huang, J.Y., Huang, M.F., Lin, S.J., Chen, C.Y., Lin, S.S., Lightener, D.V., Wang, H.C., Wang, A.H., Wang, H.C., Hor, L.I., Lo, C.F. 2015. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. PNAS 112:10798-10803.
- Liao, H.F., Mo, C.F., Wu, S.C., Cheng, D.H., Yu, C.Y., Chang, K.W., Kao, T.H., Lu, C.W., Pinskaya, M., Morillon, A., Lin, S.S., Cheng, W.T., Bourc'his, D., Bestor, T., Sung, L.Y., Lin, S.P. 2015. Dnmt3l-knockout donor cells improve somatic cell nuclear transfer reprogramming efficiency. Reproduction 150:245-256.
- Yang, C.Y., Huang, Y.H., Lin, C.P., Lin, Y.Y., Hsu, H.C., Wang, C.N., Liu, L.Y., Shen, B.N., Lin, S.S.* 2015. MicroR396-targeted SHORT VEGETATIVE PHASE is required to repress flowering and is related to the development of abnormal flower symptoms by the Phyllody Symptoms1 effector. Plant Physiol. 168:1702-1716. (*correspond author)
- Hu, S.F., Huang, Y.H., Lin, C.P., Liu, L.Y.D., Hong, S.F., Yang, C.Y., Lo, H.F., Tseng, T.Y., Chen, W.Y., Lin, S.S.* 2015. Development of a mild viral expression system for gain-of-function study of phytoplasma effector in planta. PLOS ONE. 10:e0130139 (*correspond author)
- Huang, C.H., Hsiao, W.R., Huang, C.W., Chen, K.C., Lin, S.S., Chen, T.C., Raja, J.A.J., Wu, H.W., Yeh, S.D. 2015. Two novel motifs of Watermelon silver mottle virus NSs protein are responsible for RNA silencing suppression and pathogenicity. PLOS ONE. 10:e0126161
- Lin, Y.L., Ma, L.T., Lee, Y.R., Lin, S.S., Wang, S.Y., Chang, T.T., Shaw, J.F., Li, W.H., Chu, F.F. 2015. MicroRNA-like small RNAs prediction in the development of Antrodia cinnamomea. PLOS ONE. 10:e0123245
- Lin, P.C., Hu, W.C., Lee, S.C., Chen, Y.L., Lee, C.Y., Chen, Y.R., Liu, L.Y.D., Chen, P.Y., Lin. S.S.*, Chang, Y.C.* 2015. Application of an integrated omics approach for identifying host proteins that interact with Odontoglossum ringspot virus capsid protein. Mol. Plant Microbe Interact. 28:711-726 (*co-correspond author)
- Wen, C.H., Lin, S.S., Chu, F.H. 2015. Transcriptome analysis of a subtropical deciduous tree: autumn leaf senescence gene expression profile of Formosan Gum. Plant Cell Physiol. 56:163-174
- Yang, Y.T., Chen, I.T., Lee, C.T., Chen, C.Y., Lin, S.S., Hor, L.I., Tseng, T.C., Huang, Y.T., Sritunyalucksana, K., Thitamadee, S., Wang, H.C., Lo, C.F. 2014. Draft genome sequences of four strains of Vibrio parahaemolyticus, three of which cause early mortality syndrome/acute hepatopancreatic necrosis disease in shrimp in China and Thailand. Genome Announc. 2:e00816-14
- Yang, Y.T., Lee, D.Y., Wang, Y.G., Hu, J.M., Li, W.H., Leu, J.H., Chang, G.D., Ke, H.M., Kang, S.T., Lin, S.S., Kou, G.H., Lo, C.F. 2014. The genome and occlusion bodies of marine Penaeus monodon nudivirus (PmNV, also known as MBV and PemoNPV) suggest that it should be assigned to a new nudivirus genus that is distinct from the terrestrial nudiviruses. BMC Genomics 15:628
- Kung, Y.J., Lin, P.C., Yeh, S.D., Hong, S.F., Chua, N.H., Huang, Y.H., Liu, L.Y., Lin. C.P., Wu, H.W., Chen, C.C., Lin, S.S.* 2014. Genetic analyses of the FRNK motif function of Turnip mosaic virus uncover multiple and potentially interactive pathways of cross-protection. Mol. Plant Microbe Interact. 27:944-955. (*correspond author)
- Tseng, H.I., Lin, C.P., Lin, S.S.* 2014. Characterization and identification of Catharanthus roseus epigenetic-related genes that in response to peanut witches’-broom phytoplasma-mediated infection. Plant Pathol. Bull. 23:67-77 (*correspond author)
- Liu, L.Y., Tseng, H.I., Lin, C.P., Lin, Y.Y., Huang, Y.H., Huang, C.K., Chang, T.H., Lin, S.S.* 2014. High-throughput transcriptome analysis of the leafy flower transition of Catharanthus roseus induced by peanut witches’-broom phytoplasma infection. Plant Cell Physiol. 55:942-957 (* correspond author) (Cover photo)
- Youngson, N.A., Lin, P.C., Lin, S.S.* 2013. The convergence of autophagy, small RNA and the stress response. Implications for transgenerational epigenetic inheritance in plants. BioMol. Con. 5:1-8. (*correspond author)
- Chiu, M.T., Lin, C.P., Lin, P.C., Lin, S.S.* 2013. Enhancement of IgG purification by FPLC for a serological study on the Turnip mosaic virus P1 protein. Plant Pathol. Bull. 22:21-30 (* correspond author)
- Lin, Y.Y., Fang, M.M., Lin, P.C., Chiu, M.T., Liu, L.Y., Lin, C.P., Lin, S.S.* 2013. Improving initial infectivity of the Turnip mosaic virus (TuMV) infectious clone by an mini binary vector via agro-infiltration. Botanical Studies 54:22 (* correspond author)
- Wang, L.Y., Lin, S.S., Hung, T.H., Li, T.K., Lin, N.C., Shen, T.L. 2012. Multiple domains of the Tobacco mosaic virus p126 protein can independently suppress local and systemic RNA silencing. Mol. Plant Microbe Interact. 25:648-657
- Kung Y.J., Lin, S.S., Huang Y.L., Chen, T.C., Harish, S.S., Chua, N.H., Yeh, S.D. 2012 Multiple artificial microRNAs targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded RNA plant virus. Mol. Plant Pathol. 13:303-317
- Lafforgue, G., Martinez F., Sardanyes, J. de la Iglesia, F., Niu, Q.W., Lin, S.S., Sole, R.V., Chua, N.H., Daros, J., and Elena S.F. 2011. Tempo and mode of plant RNA virus escape from RNAi-mediated resistance. J. Virol. 85:9686-9695
- Wu, H.W.*, Lin, S.S.*, Chen, K.C., Yeh, S.D., Chua, N.H. 2010. Discriminating mutations of HC-Pro of Zucchini yellow mosaic virus with differential effects on small RNA pathways involved in viral pathogenicity and symptom development. Mol. Plant Microbe Interact. 23:17-28 (*co-first author)
- Lin, S.S.*, Wu, H.W.*, Elena, S.F., Chen, K.C., Niu, Q.W., Yeh, S.D., Chen, C.C., Chua, N.H. 2009. Molecular evolution of a viral non-coding sequence under the selective pressure of amiRNA-mediated silencing. PLoS Pathog. 5(2):e1000312. Doi:10.1371/journal.ppat.1000312 (*co-first author)
- Lin, S.S., Martin, R., Mongrand, S., Vandenabeels, S., Chen, K.C., Jang, I.C., Chua, N.H. 2008. RING1 E3 ligase localizes to plasma membrane lipid raft to trigger FB1-induced programmed cell death in Arabidopsis. Plant J. 56:550-561
- Lin, S.S., Henriques, R., Wu, H.W., Niu, Q.W., Yeh, S.D., and Chua, N.H. 2007. Strategies and mechanisms of plant virus resistance. Plant Biotechnol. Rep. 1:125-134 (Review)
- Lin, S.S., H.W. Wu, F.-J. Jan, Hou, R.F., and Yeh, S.D. 2007. Modifications of the helper component-protease of Zucchini yellow mosaic for generation of attenuated mutants for cross protection against severe infection. Phytopathology 97:287-296 (Cover photo)
- Chiang, C.H., Lee, C.Y., Wang, C.H., Jan, F.J., Lin, S.S., Chen, T.C., Raja, J.A.J, and Yeh, S.D. 2007. Genetic analysis of an attenuated Papaya ringspot virus strain applied for cross-protection. Eur. J. Plant Pathol. 118:333-348
- Chen, C.C., Chen, T.C., Raja, J.A., Chang, C.A., Chen, L.W., Lin, S.S., Yeh, S.D. 2007. Effectiveness and stability of heterologous proteins expressed in plants by Turnip mosaic virus vector at five different insertion sites. Virus Res. 130:210-227
- Niu, Q.W.*, Lin, S.S.*, Reyes J.L., Chen, K. C., Wu, H.W., Yeh, Y.D., and Chua, N.H. 2006. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotech. 24:1420-1428 (*co-first author)
- Zhang, X., Yuan, Y.U., Pei, Y., Lin, S.S., Tuschl, T., Patel, D.J., and Chua, N.H. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis AGO1 cleavage activity to counter plant defense. Gene Dev. 20:3255-3268
- Zhang, X., Henriques, R., Lin, S.S., Niu, Q.W., Chua, N.H. 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral-dip method. Nat. Protocol. 1:641-646.
- Hsu, C.H., Lin, S.S., Liu, F.L., Su,W.C., and Yeh, S.D. 2004. Oral administration of mite allergen expressed by zucchini yellow mosaic virus in cucurbit species downregulates allergen-induced airway inflammation and IgE synthesis. J. Allergy Clin Immunol. 113:1079-1085.
- Lin, S. S., Hou, R. F., and Yeh, S. D. 2002. Construction of in vitro and in vivo infectious transcripts of a Taiwan strain of Zucchini yellow mosaic virus. Bot. Bull. Acad. Sin. 43:261-269.
- Lee, K.C., Lin, S.S., Yeh, S.D., and Wong, S.M. 2002. Interactions between nuclear inclusion protein a (NIa) and Nuclear Inclusion Protein b (NIb) of Zucchini yellow mosaic virus and Papaya ringspot virus. Plant Prot. Bull. 11:79-86.
- Lin, S. S., Hou, R. F., and Yeh, S. D. 2001. Complete genome sequence and genetic organization of a Taiwan isolate of Zucchini yellow mosaic virus. Bot. Bull. Acad. Sin. 42: 243-250.
- Chu F. H., Chao, C. H., Peng, Y. C. Lin, S. S., Chen, C. C., Yeh, S. D. 2001. Serological and molecular characterization of Peanut chlorotic fan-spot virus, a new species of the genus Tospovirus. Phytopathology 91: 856-863.
- Lin, S.S., Hou, R.F., and Yeh, S.D. 2000. Heteroduplex mobility and sequence analyses for assessment of variability of Zucchini yellow mosaic virus. Phytopathology 90: 228-235.
- Huang, C.H., Lin, S.S., and Yeh, S.D. 2000. Sequence analysis of the coat protein gene of Zucchini yellow mosaic virus isolated from diseased woody fruit of bitter gourd. Plant Pathol. Bull. 10:11-18.
- Lin, S.S., Hou, R.F., Huang, C.H., and Yeh, S.D. 1998. Characterization of Zucchini yellow mosaic virus (ZYMV) isolates collected from Taiwan by host reactions, serology, and RT-PCR. Plant Prot. Bull. 40: 163-176.
Book
1. Lin, S.S.*, Chen, Y., Lu, M.J.* (2019) Degradome Sequencing in Plants. In: de Folter S. (eds) Plant MicroRNAs. Methods Molecular Biology, vol 1932. pp197-213. Humana Press, New York, NY. (First & co-correspond author).