Research

Home News Research Publications Members About CSY
Application of microbial rhodopsin and Biotechnology:
Our Microbial rhodopsins (M-Rho) are found in Archaea, Bacteria and some species of Eukarya and serve as light-driven ion pumps or mediate phototaxis responses in various biological systems. We previously reported an expression system using a highly expressible mutant, D94N-HmBRI (HEBR) from Haloarcula marismortui, as a leading tag to assist in the expression of membrane proteins that were otherwise difficult to express in E. coli. In this study, we show a universal strategy for the expression of two M-Rho proteins, either the same or different types, as one fusion protein with the HEBR system. One extra transmembrane domain was engineered to the C-terminal of HEBR to express another target M-Rho. The average expression yield in this new system reached a minimum of 2 mg/L culture, and the maximum absorbance of the target M-Rho remained unaltered in the fusion forms. The fusion protein showed a combined absorbance spectrum of a lone HEBR and target M-Rho. The function of the target M-Rho was not affected after examination with functional tests, including the photocycle and proton pumping activity of fusion proteins. In addition, an otherwise unstable sensory rhodopsin, HmSRM, showed the same or even improved stability under various temperatures, salt concentrations, and a wide range of pH conditions. This HEBR platform provides the possibility to construct multi-functional, stoichiometric and color-tuning fusion proteins using M-Rho from haloarchaea.
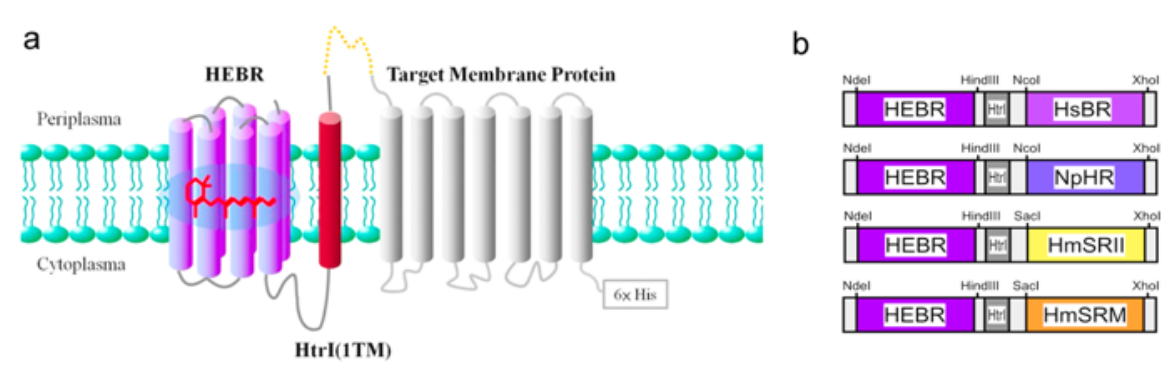
Construction strategy of the HEBR fusion with target protein(s). (a) The transmembrane region 1 (TM1) of HmHtrI was introduced between HEBR and the target membrane protein with the N-terminus residing in the periplasmic side; see the Methods for more details. (b) Diagrams summarize the tandem expression cassette variants and the designed restriction enzyme sites for the construction of the target microbial rhodopsin.
Seeking novel microbial rhodopsin system in archaea:
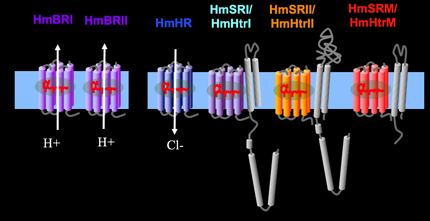
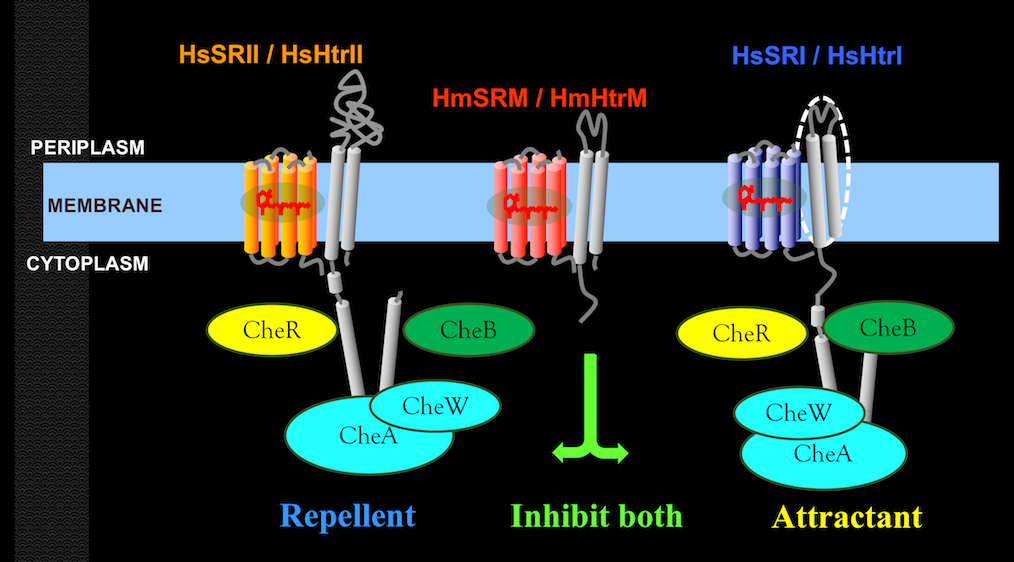
Our lab unveiled a six-rhodopsin photosensory system, the most in microbes, in Haloarcula marismortui.
The system includes three ion translocating rhodopsins, two sensory rhodopsins and one sensory-like rhodopsin.
Six-rhodopsin from Haloarcula marismortui
HmBRI: Light-driven proton outward translocator; Bacteriorhodopsins are light-driven proton translocators. Biomimicry based on photoreceptors potentially can take advantage of sun light to drive the direct production of bioenergy or indirectly. Bacteriorhodopsin, Halorhodopsin, Sensory rhodopsins, transducers and F1/Fo ATPase are the candidates.
HmBRII: Light-driven proton outward translocator; Bacteriorhodopsin-like.
HmHR: Light-driven chloride inward translocator; Halorhodopsin-like.
HmSRI: Positive phototaxis sensory rhodopsin with a congnate transducer, HmHtrI. Like those known as SRI.
HmSRII: Photo-repellent sensory rhodopsin with a cognate transducer, HmHtrII. Like those known as SRII.
HmSRM: Sensory-like rhodopsin working with a cognate transducer, HmHtrM.
Instead of containing Histidine kinase binding domain found in any transducer associated with sensory rhodopsin, HmHtrM lacks such domain.
Biological Functions of Microbial Rhodopsins and their associated proteins:
Haloarchaea utilize various microbial rhodopsins to harvest light energy or to mediate phototaxis in search of optimal environmental niches. To date, only the red light-sensing sensory rhodopsin I (SRI) and the blue light-sensing sensory rhodopsin II (SRII) have been shown to mediate positive and negative phototaxis, respectively. In this work, we demonstrated that a blue-green light-sensing (504 nm) sensory rhodopsin from Haloarcula marismortui, SRM, attenuated both positive and negative phototaxis through its sensing region. The H. marismortui genome encodes three sensory rhodopsins: SRI, SRII and SRM. Using spectroscopic assays, we first demonstrated the interaction between SRM and its cognate transducer, HtrM. We then transformed an SRM-HtrM fusion protein into Halobacterium salinarum, which contains only SRI and SRII, and observed that SRM-HtrM fusion protein decreased both positive and negative phototaxis of H. salinarum. Together, our results suggested a novel phototaxis signalling system in H. marismortui comprised of three sensory rhodopsins in which the phototactic response of SRI and SRII were attenuated by SRM.
Structural study on protein science:
Structural studies facilitate our understanding of the molecular mechanisms mediating functions.
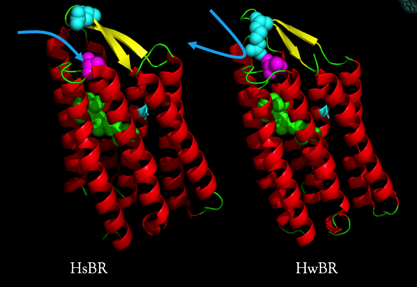
We resolved the crystal structure of an acid-tolerance bacteriorhodopsin from Haloquadratum walsbyi and published it at 2015.
G-protein inteaction with other small chemicals, oligo peptides, and proteins:
Our lab is also working on G-protein related pathways, including GPCR, RGS and G-protein signaling: basic scientific facts and biotechnological studies
A G-protein related screening system for small molecules and/or proteins is under intense development.